# @markdown #Run this cell only once per notebook instance.
# @markdown This cell is responsible for installing the Paddle base framework
# and the segmentation version called PaddleSeg. After the installation is
# complete a test script will run, it will download a small dataset and run
# a neural network for a few iterations to verify that everything was installed
# successfully.
# @markdown You can check the box below to download and extract the cloud
# dataset. This dataset contains 1223 images that contain the following 6
# classes: Sky, Tree, Stratocumuliform, Stratiform, Cirriform and Cumuliform.
# This dataset is already in the expected format, split into training and
# validation, and can be used as a reference to adapt your dataset to the
# expected format for PaddleSeg. At the end of the notebook we have a simple
# script to convert a dataset to PaddleSeg's format.
# @markdown ----
download_cloud_dataset = True # @param {type:"boolean"}
# @markdown ----
# @markdown  # noqa: E501
!pip install paddlepaddle-gpu
import paddle
from google.colab import drive
paddle.utils.run_check()
print(paddle.__version__)
!git clone https://github.com/PaddlePaddle/PaddleSeg
%cd PaddleSeg
!pip install -r requirements.txt
!sh tests/run_check_install.sh
!python setup.py install
drive.mount("/content/drive")
%cd '/content/'
if download_cloud_dataset:
!gdown 1nuk9mBOAQgaPF9WxnoKDBtGXFh3cUeEH
!unzip '/content/PaddleSegNuvens-ComArvore-1223.zip'PaddleSeg generic example
In this tutorial you can change the model choice between: OCRNet, SegFormer and PPLiteSeg.
Setup Inicial
Opções Gerais
# @markdown #Dataset settings.
# @markdown Enter a file path:
dataset_root = "/content/PaddleSegNuvens-ComArvore-1223" # @param {type:"string"}
folder_name_dataset = dataset_root.split("/")[-1]
train_path = (
"/content/PaddleSegNuvens-ComArvore-1223/train-paddle.txt" # @param {type:"string"}
)
val_path = (
"/content/PaddleSegNuvens-ComArvore-1223/val-paddle.txt" # @param {type:"string"}
)
num_classes = 6 # @param {type:"number"}
# @markdown #Mean and standard deviation of the dataset, where r = red,
# g = green and b = blue.
mean_r = 0.37555224 # @param {type:"number"}
mean_g = 0.47573688 # @param {type:"number"}
mean_b = 0.51197395 # @param {type:"number"}
std_r = 0.37555224 # @param {type:"number"}
std_g = 0.47573688 # @param {type:"number"}
std_b = 0.51197395 # @param {type:"number"}
# @markdown #Train settings.
batch_size = 4 # @param {type:"number"}
iters = 80000 # @param {type:"number"}
# @markdown #Enter the desired size so that the image will be resized to this
# value. Set the original size so that it does not resize.
target_size_x = 512 # @param {type:"number"}
target_size_y = 512 # @param {type:"number"}
size_folder_name = f"{target_size_x}-{target_size_y}"
# @markdown #Transforms. These values add up to both the up and down
# transformation. So 10 saturation will take the original value and can add or
# remove up to 10 saturation units.
saturation_range = 0.5 # @param {type:"slider", min:0, max:1, step:0.05}
contrast_range = 0.20 # @param {type:"slider", min:0, max:1, step:0.1}
brightness_range = 0.20 # @param {type:"slider", min:0, max:1, step:0.1}
base = f"""
batch_size: {batch_size}
iters: {iters}
train_dataset:
type: Dataset
dataset_root: {dataset_root}
train_path: {train_path}
num_classes: {num_classes}
transforms:
- type: Resize
target_size: [{target_size_x}, {target_size_y}]
- type: RandomHorizontalFlip
- type: RandomVerticalFlip
- type: RandomDistort
brightness_range: {brightness_range}
contrast_range: {contrast_range}
saturation_range: {saturation_range}
- type: Normalize
mean: [{mean_r}, {mean_g}, {mean_b}]
std: [{std_r}, {std_g}, {std_b}]
mode: train
val_dataset:
type: Dataset
dataset_root: {dataset_root}
val_path: {val_path}
num_classes: {num_classes}
transforms:
- type: Resize
target_size: [{target_size_x}, {target_size_y}]
- type: Normalize
mean: [{mean_r}, {mean_g}, {mean_b}]
std: [{std_r}, {std_g}, {std_b}]
mode: val
"""Models
Run only one of the cells in this section. If you want to change the experiment, modify and run the cell again, or choose another cell.
OCRNet
# @markdown #HRNet
# @markdown ---
# @markdown #Network Size
HrNetSize = "48" # @param ["18", "48"]
# @markdown ---
# @markdown #Paper: https://arxiv.org/abs/1909.11065
# @markdown #Github: https://github.com/PaddlePaddle/PaddleSeg/tree/release/2.6/configs/ocrnet # noqa: E501
# @markdown #Overview We propose a high-resolution network (HRNet). The HRNet
# maintains high-resolution representations by connecting high-to-low
# resolution convolutions in parallel and strengthens high-resolution
# representations by repeatedly performing multi-scale fusions across
# parallel convolutions. We demonstrate the effectives on pixel-level
# classification, region-level classification, and image-level classification.
# @markdown # noqa: E501 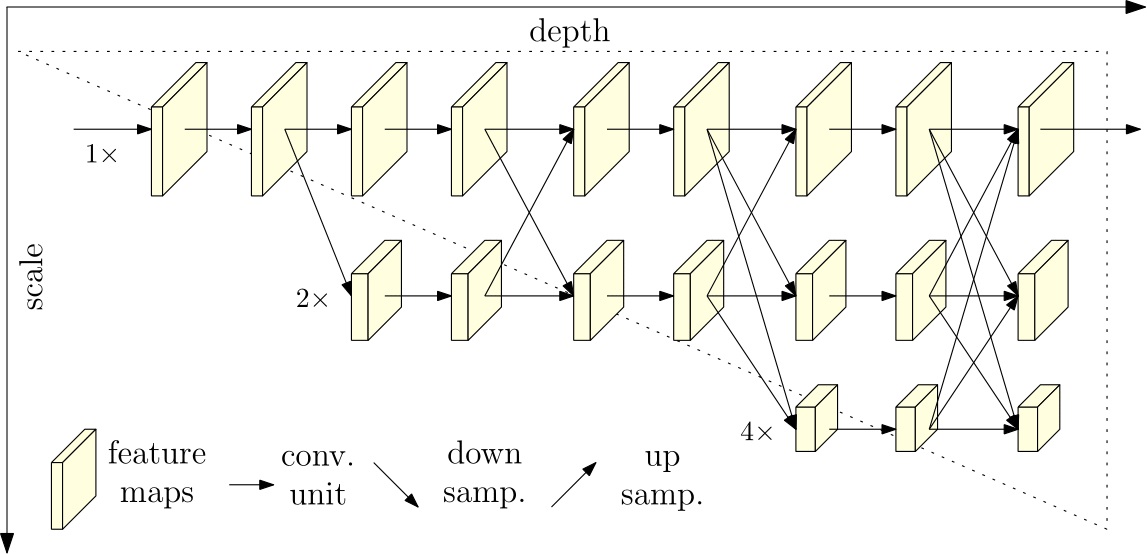
model_folder_name = f"HrNet-{HrNetSize}"
logits_size = 2
model = f"""
model:
type: OCRNet
backbone:
type: HRNet_W{HrNetSize}
pretrained: https://bj.bcebos.com/paddleseg/dygraph/hrnet_w{HrNetSize}_ssld.tar.gz
backbone_indices: [0]
""" # noqa: E501SegFormer
# @markdown #SegFormer
# @markdown ---
# @markdown #Network size.
model_depth = "B3" # @param ["B1", "B2", "B3", "B4", "B5"]
# @markdown ---
# @markdown #PPLiteSeg
# @markdown #Paper: https://arxiv.org/abs/2105.15203
# @markdown #Github: https://github.com/PaddlePaddle/PaddleSeg/tree/release/2.6/configs/segformer # noqa: E501
# @markdown #Overview:
# @markdown SegFormer is a Transformer-based framework for semantic
# segmentation that unifies Transformers with lightweight multilayer
# perceptron (MLP) decoders. SegFormer has two appealing features:
# 1) SegFormer comprises a novel hierarchically structured Transformer
# encoder which outputs multiscale features. It does not need positional
# encoding, thereby avoiding the interpolation of positional codes which leads
# to decreased performance when the testing resolution differs from training.
# 2) SegFormer avoids complex decoders. The proposed MLP decoder aggregates
# information from different layers, and thus combining both local attention
# and global attention to render powerful representations.
# @markdown  # noqa: E501
model_depth_down = model_depth.lower()
model_folder_name = f"SegFormer-{model_depth}"
logits_size = 2
model = f"""
model:
type: SegFormer_{model_depth}
num_classes: {num_classes}
pretrained: https://bj.bcebos.com/paddleseg/dygraph/mix_vision_transformer_{model_depth_down}.tar.gz
"""PPLiteSeg
# @markdown #PPLiteSeg
# @markdown ---
# @markdown #Network size.
STDC = 2 # @param {type:"slider", min:1, max:2, step:1}
# @markdown ---
# @markdown #Paper: https://arxiv.org/abs/2204.02681
# @markdown #Github: https://github.com/PaddlePaddle/PaddleSeg/tree/release/2.6/configs/pp_liteseg # noqa: E501
# @markdown #Overview:
# @markdown Overview: We propose PP-LiteSeg, a novel lightweight model for the
# real-time semantic segmentation task. Specifically, we present a Flexible and
# Lightweight Decoder (FLD) to reduce computation overhead of previous decoder.
# To strengthen feature representations, we propose a Unified Attention Fusion
# Module (UAFM), which takes advantage of spatial and channel attention to
# produce a weight and then fuses the input features with the weight. Moreover,
# a Simple Pyramid Pooling Module (SPPM) is proposed to aggregate global
# context with low computation cost.
# @markdown  # noqa: E501
model_folder_name = f"PPLiteSeg-{STDC}"
logits_size = 3
model = f"""
model:
type: PPLiteSeg
backbone:
type: STDC{STDC}
pretrained: https://bj.bcebos.com/paddleseg/dygraph/PP_STDCNet{STDC}.tar.gz
arm_out_chs: [32, 64, 128]
seg_head_inter_chs: [32, 64, 64]
"""Optimizer
If you want to change the experiment, modify and run the cell again, or choose another cell.
AdamW
# @markdown #AdamW
# @markdown #Paper: https://arxiv.org/pdf/1711.05101.pdf
# @markdown #API: https://www.paddlepaddle.org.cn/documentation/docs/en/2.2/api/paddle/optimizer/AdamW_en.html#adamw # noqa: E501
# @markdown #The exponential decay rate for the 1st moment estimates.
beta1 = 0.4 # @param {type:"slider", min:0, max:1, step:0.1}
# @markdown #The exponential decay rate for the 2nd moment estimates.
beta2 = 0.984 # @param {type:"slider", min:0, max:1, step:0.001}
# @markdown #The weight decay coefficient.
weight_decay = 0.001 # @param {type:"number"}
optimizer = f"""
optimizer:
type: AdamW
beta1: {beta1}
beta2: {beta2}
weight_decay: {weight_decay}
"""SGD
# @markdown #SGD
# @markdown #API: https://www.paddlepaddle.org.cn/documentation/docs/en/2.2/api/paddle/optimizer/SGD_en.html#sgd # noqa: E501
momentum = 0.9 # @param {type:"slider", min:0, max:1, step:0.1}
weight_decay = 0.0005 # @param {type:"number"}
optimizer = f"""
optimizer:
type: sgd
momentum: {momentum}
weight_decay: {weight_decay}
"""Learning Rate
If you want to change the experiment, modify and run the cell again, or choose another cell.
PolynomialDecay
# @markdown #PolynomialDecay
learning_rate = 0.9 # @param {type:"number"}
weight_decay = 0.0005 # @param {type:"number"}
warmup_iters = 1000 # @param {type:"number"}
warmup_start_lr_power = 5 # @param {type:"slider", min:0, max:6, step:1}
lr_scheduler = f"""
lr_scheduler:
type: PolynomialDecay
learning_rate: {learning_rate}
end_lr: 0
power: 0.9
warmup_iters: {warmup_iters}
warmup_start_lr: 1.0e-{warmup_start_lr_power}
"""Loss
If you want to change the experiment, modify and run the cell again, or choose another cell.
CrossEntropyLoss
# @markdown #CrossEntropyLoss
# @markdown #Api: https://github.com/PaddlePaddle/PaddleSeg/blob/release/2.6/paddleseg/models/losses/cross_entropy_loss.py # noqa: E501
# weight_list = '0.9, 1.0, 1.0, 1.0, 1.0, 1.0, 1.2' #@param {type:"string"}
folder_name_loss = "CrossEntropyLoss"
loss = f"""
loss:
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
coef: [1]
coef: {[1 for i in list(range(logits_size))]}
"""DetailAggregateLoss (Single Class)
# @markdown #DetailAggregateLoss (Single Class)
# @markdown #Paper: https://arxiv.org/abs/2104.13188
# @markdown #API: https://github.com/PaddlePaddle/PaddleSeg/blob/release/2.6/paddleseg/models/losses/detail_aggregate_loss.py # noqa: E501
folder_name_loss = "DetailAggregateLoss"
cross_entropy_weight = 0.2 # @param {type:"slider", min:0, max:1, step:0.1}
detail_aggregated_weight = 0.2 # @param {type:"slider", min:0, max:1, step:0.1}
loss = f"""
loss:
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
- type: DetailAggregateLoss
coef: [{cross_entropy_weight}, {detail_aggregated_weight}]
coef: {[1 for i in list(range(logits_size))]}
"""EdgeAttentionLoss (Single Class)
# @markdown #EdgeAttentionLoss (Single Class)
# @markdown #Implements the cross entropy loss function. It only compute the edge part.
# @markdown #API: https://github.com/PaddlePaddle/PaddleSeg/blob/release/2.6/paddleseg/models/losses/edge_attention_loss.py
folder_name_loss = "EdgeAttentionLoss"
cross_entropy_weight = 0.8 # @param {type:"slider", min:0, max:1, step:0.1}
edge_attention_weight = 0.2 # @param {type:"slider", min:0, max:1, step:0.1}
loss = f"""
loss:
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
- type: EdgeAttentionLoss
coef: [{cross_entropy_weight}, {edge_attention_weight}]
coef: {[1 for i in list(range(logits_size))]}
"""PixelContrastCrossEntropyLoss (Arrumar)
# @markdown #PixelContrastCrossEntropyLoss (Arrumar)
# @markdown #Paper: https://arxiv.org/abs/2101.11939
# @markdown #API: https://github.com/PaddlePaddle/PaddleSeg/blob/release/2.6/paddleseg/models/losses/pixel_contrast_cross_entropy_loss.py # noqa: E501
folder_name_loss = "PixelContrastCrossEntropyLoss"
temperature = 0.1 # @param {type:"number"}
base_temperature = 0.07 # @param {type:"number"}
max_samples = 1024 # @param {type:"number"}
max_views = 100 # @param {type:"number"}
cross_entropy_weight = 0.8 # @param {type:"slider", min:0, max:1, step:0.1}
pixel_contrast_weight = 0.2 # @param {type:"slider", min:0, max:1, step:0.1}
loss = f"""
loss:
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
- type: PixelContrastCrossEntropyLoss
temperature: {temperature}
base_temperature: {base_temperature}
ignore_index: 255
max_samples: {max_samples}
max_views: {max_views}
coef: [{cross_entropy_weight}, {pixel_contrast_weight}]
coef: {[1 for i in list(range(logits_size))]}
"""SemanticConnectivityLoss
# @markdown #SemanticConnectivityLoss
# @markdown #Paper: https://arxiv.org/abs/2112.07146
# @markdown #API: https://github.com/PaddlePaddle/PaddleSeg/blob/release/2.6/paddleseg/models/losses/semantic_connectivity_loss.py # noqa: E501
folder_name_loss = "SemanticConnectivityLoss"
# @markdown Maximum number of predicted connected components. At the beginning
# of training, there will be a large number of connected components, and the
# calculation is very time-consuming. Therefore, it is necessary to limit the
# maximum number of predicted connected components, and the rest will not
# participate in the calculation.
max_pred_num_conn = 10 # @param {type:"number"}
cross_entropy_weight = 0.8 # @param {type:"slider", min:0, max:1, step:0.1}
semantic_connectivity_weight = 0.2 # @param {type:"slider", min:0, max:1, step:0.1}
loss = f"""
loss:
types:
- type: MixedLoss
losses:
- type: CrossEntropyLoss
- type: SemanticConnectivityLoss
max_pred_num_conn: {max_pred_num_conn}
coef: [{cross_entropy_weight}, {edge_attention_weight}]
coef: {[1 for i in list(range(logits_size))]}
"""Run Experiment
!export CUDA_VISIBLE_DEVICES=0
#@title Run Experiment
import os
file_content = base + model + optimizer + loss + lr_scheduler
experiment_name = os.path.join(folder_name_dataset, model_folder_name, folder_name_loss)
resume_model = False
save_interval = 200 #@param {type:"number"}
save_dir_path = '/content/drive/Shareddrives/Nuvens/0Allan/PaddleSegTest' #@param {type:"string"}
save_dir_exp = os.path.join(save_dir_path, experiment_name)
config_file = os.path.join(save_dir_exp, 'config-file.yml')
os.makedirs(save_dir_exp, exist_ok=True)
#@markdown Resume Experiment?
resume_experiment = False #@param {type:"boolean"}
checkpoint_path = "/content/drive/Shareddrives/Nuvens/resultados_allan/allan/paddleseg/hrnet18-ocr-comarvore-halfres/iter_9500" #@param {type:"string"}
resume_config_file = '/content/drive/Shareddrives/Nuvens/0Allan/PaddleSegTest/PaddleSegNuvens-ComArvore-1223/HrNet-48/DetailAggregateLoss/config-file.yml' #@param {type:"string"}
with open(config_file, "w") as text_file:
text_file.write(file_content)
print(config_file)
!python /content/PaddleSeg/train.py \
--config $config_file \
--do_eval \
--use_vdl \
--save_interval $save_interval \
--save_dir $save_dir_exp
if resume_experiment:
print(f'Resuming from {save_dir_exp}')
!python /content/PaddleSeg/train.py \
--config $resume_config_file \
--do_eval \
--use_vdl \
--save_interval $save_interval \
--save_dir $save_dir_exp \
--resume_model $checkpoint_path
#@title Predict Folder with trained model
#@markdown Files created in the training Experiment
checkpoint_path = "/content/drive/Shareddrives/Nuvens/resultados_allan/allan/paddleseg/hrnet18-ocr-comarvore-halfres/iter_9500" #@param {type:"string"}
config_file = '/content/drive/Shareddrives/Nuvens/0Allan/PaddleSegTest/PaddleSegNuvens-ComArvore-1223/HrNet-48/DetailAggregateLoss/config-file.yml' #@param {type:"string"}
model_params = '/content/drive/Shareddrives/Nuvens/resultados_allan/allan/paddleseg/hrnet18-ocr-comarvore-halfres/best_model/model.pdparams' #@param {type:"string"}
#@markdown Folder to predict
image_folder = '/content/drive/MyDrive/Datasets/2022-05-13' #@param {type:"string"}
#@markdown Folder to save the predictions
dest_folder = '/content/drive/Shareddrives/Nuvens/resultados_allan/allan/paddleseg/hrnet18-ocr-comarvore-halfres/cam1-2022-05-13' #@param {type:"string"}
#@markdown Custom color pallet, the format is a sequential RGB value for each class, and all values are separated by a space.
#@markdown In the example bellow, 0 0 0 is the value for the class zero, 7 25 163 is the value for the class one and so and on.
color_pallet = '0 0 0 7 25 163 20 85 189 32 145 215 45 205 241 42 255 49' #@param {type:"string"}
!python /content/PaddleSeg/predict.py \
--config $config_file \
--model_path \
--image_path $image_folder \
--save_dir $dest_folder \
--custom_color $color_pallet# @title String fields
import glob
import os
from pathlib import Path
from sklearn.model_selection import train_test_split
from tqdm import tqdm
mask_ext = "png" # @param {type:"string"}
image_ext = "jpg" # @param {type:"string"}
dataset_root = "/content/PaddleSegNuvens-ComArvore-1223/" # @param {type:"string"}
masks = glob.glob(
os.path.join(Path(dataset_root), "**", f"*.{mask_ext}"), recursive=True
)
mask_image_tuple_list = []
print(f"Number of masks found: {len(masks)}")
for mask in tqdm(masks):
file_name = Path(mask).stem
image = glob.glob(
os.path.join(Path(dataset_root), "**", f"{file_name}.{image_ext}"),
recursive=True,
)[0]
mask_relative = mask.replace(dataset_root, "")
image_relative = image.replace(dataset_root, "")
mask_image_tuple_list.append((image_relative, mask_relative))
validation_percentage = 0.25 # @param {type:"slider", min:0.1, max:0.9, step:0.05}
X_train, X_test = train_test_split(mask_image_tuple_list, test_size=0.2)
train_file = os.path.join(dataset_root, "train-paddle2.txt")
val_file = os.path.join(dataset_root, "val-paddle2.txt")
with open(train_file, "w") as file:
print(f"Train Size: {len(X_train)}")
for line in tqdm(X_train):
file.write(f"{line[0]} {line[1]}\n")
with open(val_file, "w") as file:
print(f"Validation Size: {len(X_test)}")
for line in tqdm(X_test):
file.write(f"{line[0]} {line[1]}\n")